







|
|
cDNA Image Denoising and Enhancement
To reduce, if not eliminate, noise and impairments present in
the acquired microarray image, thus reducing processing errors from
propagating further down the processing pipeline to the essential gene
expression analysis, image processing operations, such as denoising and
enhancement, data normalization, and spot localization and segmentation, are
routinely utilized.
Since microarray images are nonlinear in nature, nonlinear
methods can potentially preserve important structural elements such as spot
edges, and at the same time eliminate microarray image impairments. Given
the fact that cDNA microarray images are represented as a two-dimensional
array of two-component cDNA vectors, the appropriate cDNA microarray
processing solution should integrate well-known concepts from the areas of
nonlinear filtering, multidimensional scaling, and robust estimation theory.
| |
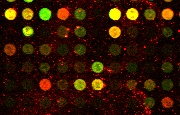
Acquired cDNA image |
|

Denoised/enhanced image |
|
|
The most commonly used method to decrease the level
of random noise present in the signal is smoothing. Therefore, a
low-pass filtering operation is required in order to replace the
noisy cDNA vectors with suitable cDNA vector representatives for the
local microarray image area determined by the supporting window. One
of the most natural approaches of identifying the atypical, noisy
samples in the data population is the sample ordering which can be
used to determine the positions of the different input vectors
without any prior information regarding the signal distributions.
Thus, vector order-statistics filters are considered to be robust
estimators and have been successfully used in cDNA microarray image
processing. Alternatively, fuzzy vector filters are highly
appropriate for microarray image denoising and enhancement because
they are capable to overcome non-stationarity in edges and can
distinguish between noise and edge pixels. |
|
References: |
|
 | R. Lukac and K.N. Plataniotis, "cDNA Microarray
Image Segmentation Using Root Signals," International
Journal of Imaging Systems and Technology, vol. 16, no. 2,
pp. 51-64, April 2006. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "cDNA Microarray Image Processing Using Fuzzy
Vector Filtering Framework," Fuzzy Sets and Systems,
Special Issue on Fuzzy Sets and Systems in Bioinformatics,
vol. 152, no. 1, pp.17-35, May 2005. |
 | R. Lukac, B. Smolka, K. Martin, K.N.
Plataniotis, and A.N. Venetsanopoulos, "Vector Filtering for Color
Imaging," IEEE Signal Processing Magazine, Special
Issue on Color Image Processing, vol. 22, no. 1, pp. 74-86, January 2005. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "A Multichannel Order-Statistic Technique for
cDNA Microarray Image Processing," IEEE Transactions on NanoBioscience,
vol. 3, no. 4, pp. 272-285, December 2004. |
|
|
|
