







|
|
cDNA Microarray Characteristics
To effectively process microarray
images, the differences among cDNA vectors spawned by the supporting
window of the image processing solution have to be quantified. Since
each cDNA vector is uniquely determined by its magnitude and direction
in the two-component RG space, the microarray image processing operators
performing data normalization, image denoising and enhancement, and spot
localization and segmentation should take into consideration both the
magnitude and the orientation of the cDNA samples.
| |

Ideal cDNA image |
Under the ideal conditions, each spot
has its own unique magnitude and directional characteristics.
Unfortunately, due to the nature of cDNA microarray technology the ideal
noise free-data are unavailable in cDNA microarray imaging, and the
acquired cDNA vectors differ in both the magnitude and the direction. |
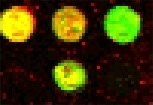
Real cDNA image |
|
| |
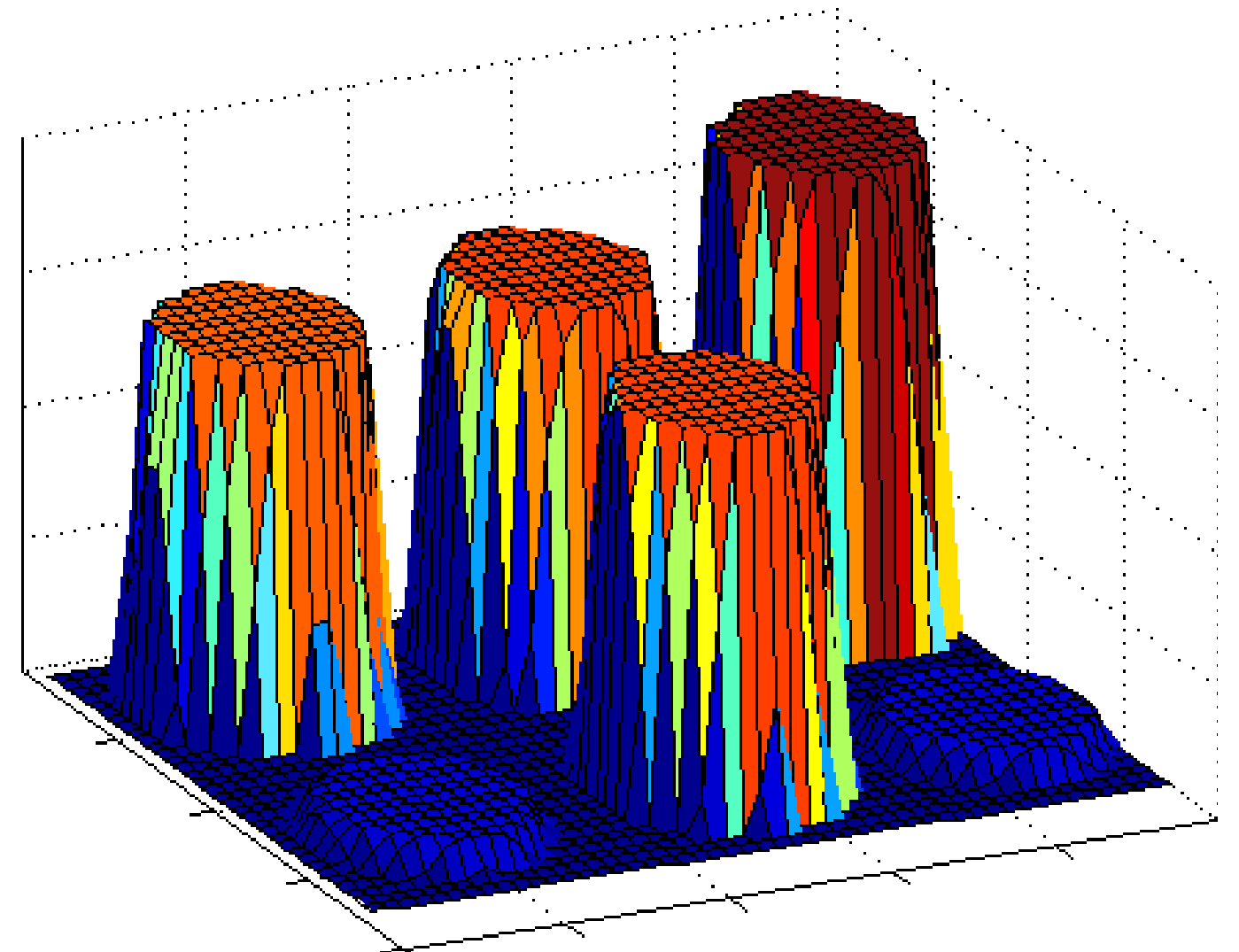
Magnitude characteristics of the ideal cDNA image |
|
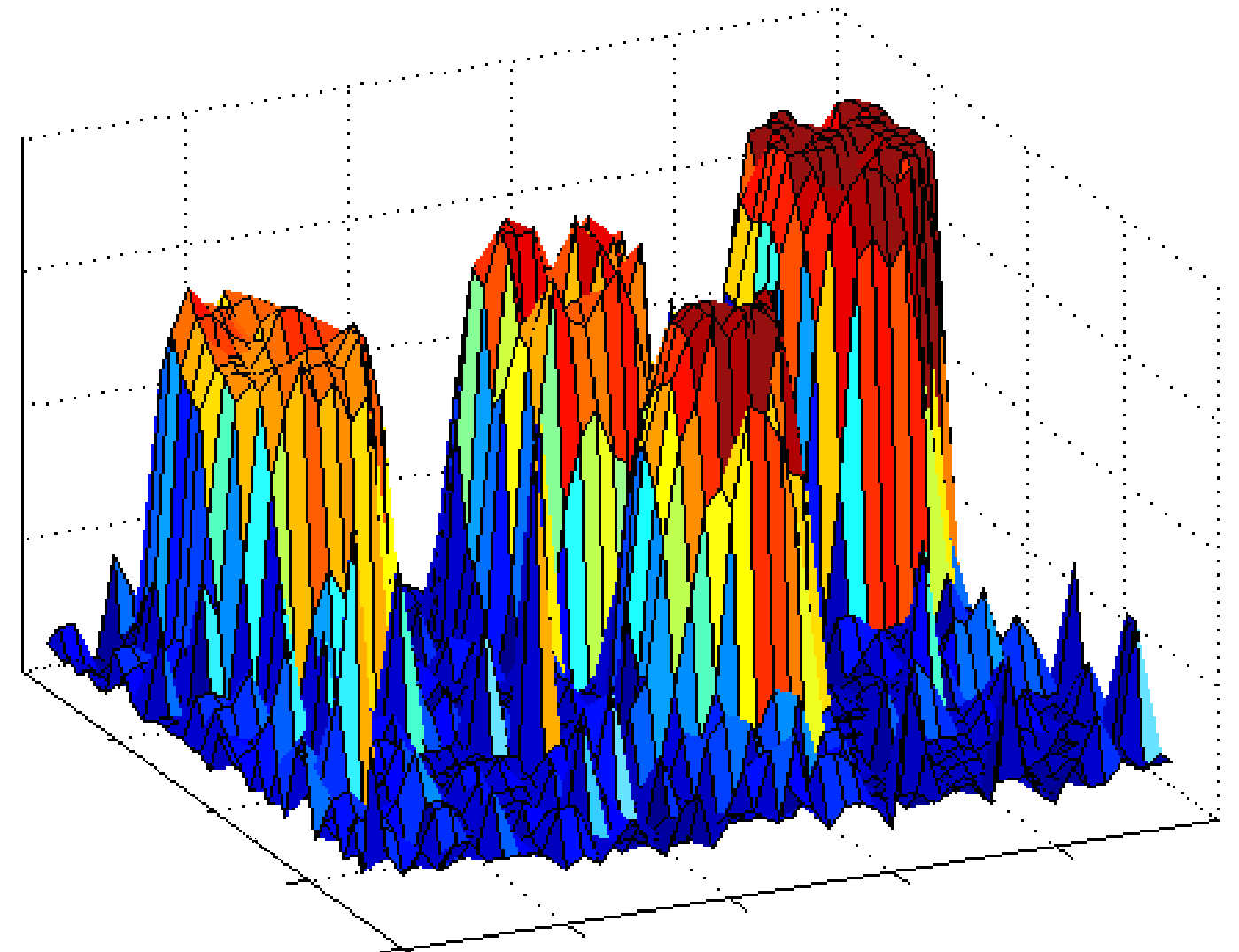
Magnitude characteristics of the real cDNA image |
|
| |
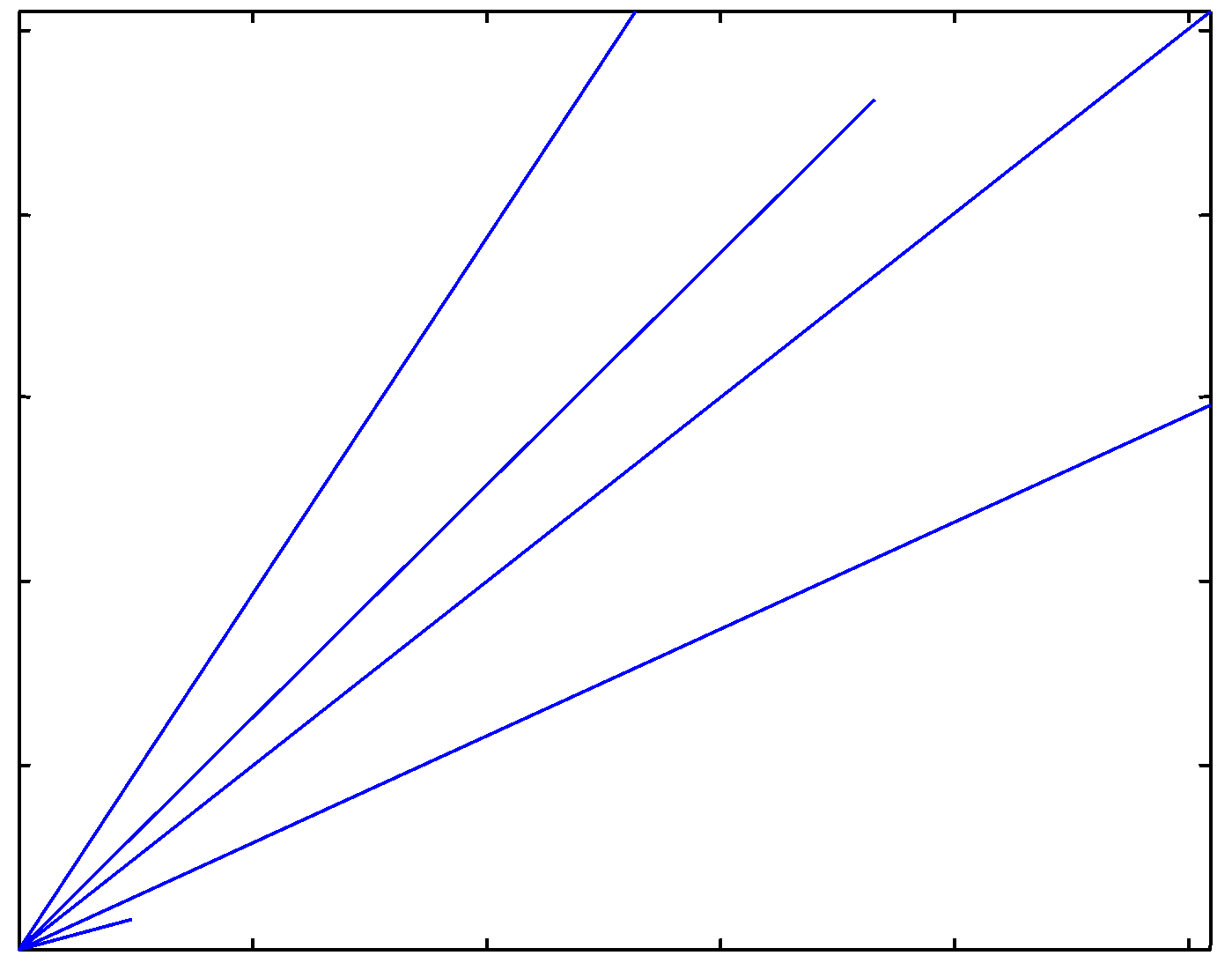
Directional characteristics of the ideal cDNA image |
|
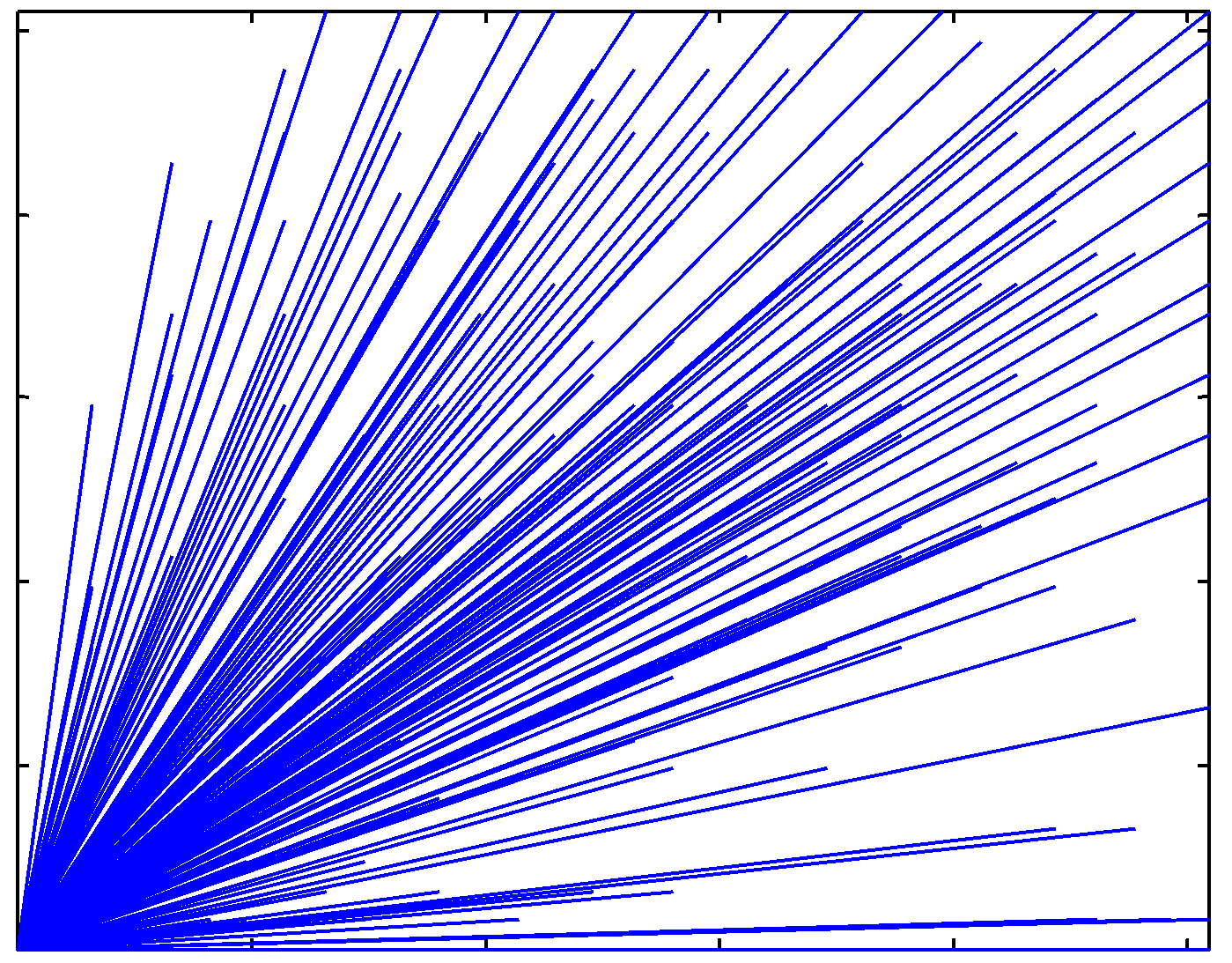
Directional characteristics of the real cDNA image |
|
|
References: |
|
 | R. Lukac and K.N. Plataniotis, "cDNA Microarray
Image Segmentation Using Root Signals," International
Journal of Imaging Systems and Technology, vol. 16, no. 2,
pp. 51-64, April 2006. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "cDNA Microarray Image Processing Using Fuzzy
Vector Filtering Framework," Fuzzy Sets and Systems, Special Issue on Fuzzy Sets and Systems in Bioinformatics,
vol. 152, no. 1, pp.17-35, May 2005. |
 | R. Lukac, B. Smolka, K. Martin, K.N.
Plataniotis, and A.N. Venetsanopoulos, "Vector Filtering for Color
Imaging," IEEE Signal Processing Magazine, Special
Issue on Color Image Processing, vol. 22, no. 1, pp. 74-86, January 2005. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "A Multichannel Order-Statistic Technique for
cDNA Microarray Image Processing," IEEE Transactions on NanoBioscience,
vol. 3, no. 4, pp. 272-285, December 2004. |
|
|
|
